
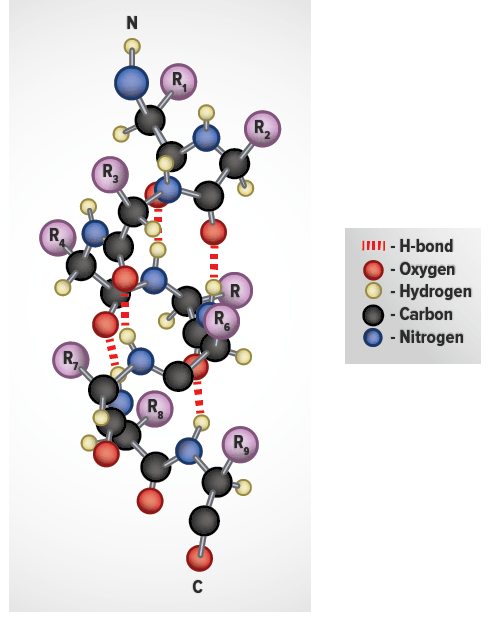
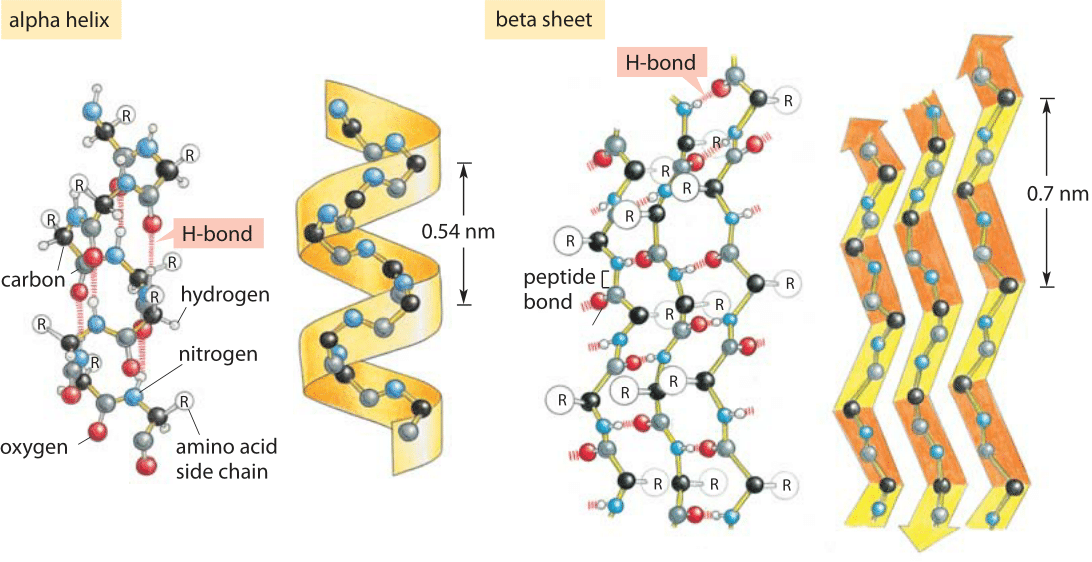
Most of the proteins for which structures has been determined have secondary structures consisting largely of alpha helices and/or beta-sheets which are tighly packed together to bury the hydrophobic amino acids (typically aliphatic and aromatic amino acids) away from the solvent. For example, many hormone receptor proteins, or large portions of them, are natively unfolded. Some proteins, often those with a very high percentage of hydrophilic amino acids, are natively unfolded, or are said to have a random coil structure. Hydrogen bonding also plays a significant role in a protein's structure, especially in regards to the secondary structure elements. This is accomplished by both local secondary structure and by the overall tertiary structure of the protein. Proteins typically fold up in such as way to minimize the solvent exposure of the hydrophobic amino acids while exposing the hydrophilic ones. To a large degree the shape of a protein is determined by the hydrophobic amino acids which prefer not to be solvated and by the hydrophilic amino acids, which like to be exposed to water. 8 Methods used to determine protein structureĪ Protein is a linear polymer of different amino acids, whose properties vary.The following section will examine how secondary structure elements connect, forming common motifs, folds, and domains. During sequence alignment and modeling, when it is essential to have an accurate sequence alignment, the highly variable length of loops justifies the localization of insertions and deletions in the amino acid sequence to loop regions. It presumably gives a protein additional stability at high temperatures, preventing its unfolding and denaturation. Loop length in organisms living at elevated temperatures (thermophilic organisms) is usually shorter than in proteins from lower-temperature family members. Nevertheless, sometimes, when a loop has some specific function, for example, interaction with another protein, the sequence may be conserved. The amino acid sequences in loop regions are often highly variable within a protein family. Short turns and longer loops are essential in protein 3D structures, connecting strands to strands, strands to α-helices, or helices to helices. The loop between the two strands is called a β-turn. When there are only two antiparallel β-strands, like in the Figure on the right, the structural motif is called a β-hairpin. The hydrogen bonds are shown as dashed lines. Together these groups form a hydrogen bond, one of the main forces in stabilizing secondary structures in proteins. When looking at the helix in the Figure below, notice how the carbonyl (C=O) oxygen atoms (shown in red) point in one direction towards the amide NH groups four residues away (i, i+4). This means that the particular configuration of the polypeptide chain is such that main chain atoms are packed in an optimal and energetically favorable way avoiding clashes (atoms coming too close to each other). In the section on the Ramachandran plot, we call these regions "energetically most favorable". This is reflected in the clustering of the torsion angles within the helical region of the Ramachandran plot. The repeating structural pattern in helices results from similar φ and ψ values. Each residue is translated 1.5 Å along the helix axis, which gives a vertical distance of 5.4 Å between structurally equivalent atoms in a turn (pitch of a turn). There are 3.6 residues/turn in an α-helix, which means that there is one residue every 100 degrees of rotation (360/3.6). In the image, only the main chain atoms of the polypeptide are shown connected by sticks, and the side chains are omitted. This type of protein structure representation is called "stick representation". An example of an α-helix is shown in the image below. The prediction was confirmed when the first three-dimensional structure of a protein, myoglobin (by Max Perutz and John Kendrew) was determined by X-ray crystallography. Linus Pauling was the first to predict the existence of α-helices. The most common type of secondary structure in proteins is the α-helix.


 0 kommentar(er)
0 kommentar(er)
